Project: Physics Informed AI for improved cancer prognosis
Description
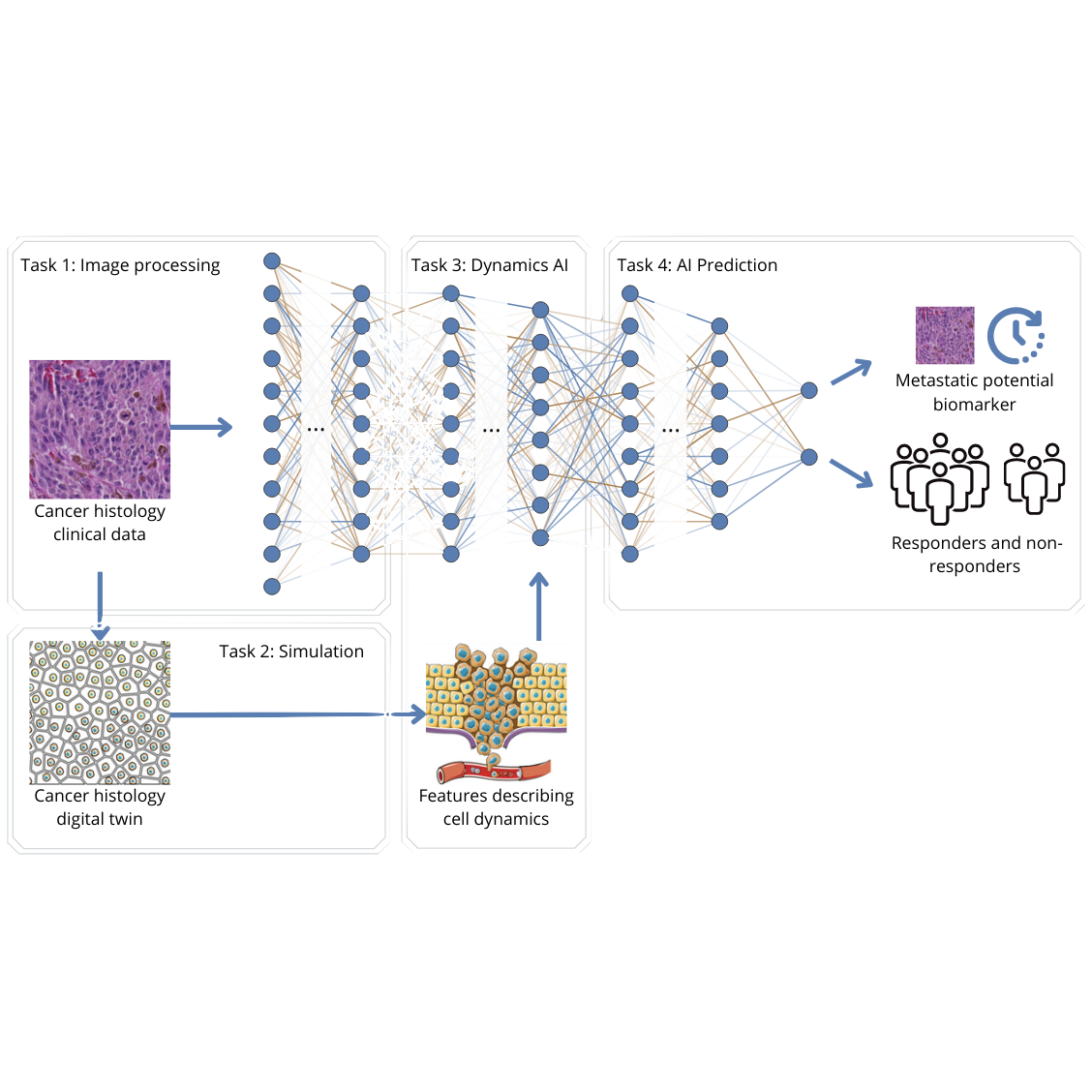 In order to metastasize, cancer cells need to move. Estimating the ability for cells to move, i.e. their dynamics, or so-called migration potential, is a promising new indicator for cancer patient prognosis (overall survival) and response to therapy. However, predicting the migration potential from static microscopic images that are routinely used in clinical practice remains a major open challenge.
In order to metastasize, cancer cells need to move. Estimating the ability for cells to move, i.e. their dynamics, or so-called migration potential, is a promising new indicator for cancer patient prognosis (overall survival) and response to therapy. However, predicting the migration potential from static microscopic images that are routinely used in clinical practice remains a major open challenge. In other words, to make a patient prognosis based on the patient's cell dynamics, we need to learn how a static microscopic image of cells can be translated to a "video", that displays the dynamics of the cells. Those videos can be generated by means of the Cellular Potts model, which simulate the physics of cell movements. However, those models need as input a set of parameters, that describe the dynamics. The goal is now, to learn by means of machine learning a model that is able to predict these parameters. In a first master thesis, we have generated a proof of concept, where a single parameter (cell volume) is learned for a single cell. However, this model needs to be extended to more parameters and multiple cells.
This topic might be for you if you like to work on an impactful application, you like to code (Pytorch), and to think creatively. The model developed in the first master thesis basically recontextualizes concepts from traditional neural networks (such as convolution) to gain realistic cell representations during the simulation of cell movement. A challenge of this project is to define architectures that are suitable to reflect Cellular Potts simulations based on Langevin dynamics. Hence, a knowledge about design choices in neural networks and their effects, or at least an interest in them is desirable.
Details
- Student
-
BMBram Meijer
- Supervisor
-
 Sibylle Hess
Sibylle Hess
- Secondary supervisor
-
SVSecondary supervisors could be Liesbeth Janssen or Mitko Vetka.